Online Analysis#
Being able to visualize and interpret simulation data in real time is invaluable for understanding the behavior of a physical system.
SmartSim can be used to stream data from Fortran, C, and C++ simulations to Python where visualization is significantly easier and more interactive.
This example shows how to use SmartSim analyze the vorticity field during a simple, Python based Lattice Boltzmann fluid flow simulation.
Lattice Boltzmann Simulation#
This example was adapted from Philip Mocz’s implementation of the lattice Boltzmann method in Python. Since that example is licensed under GPL, so is this example.
Philip also wrote a great medium article explaining the simulation in detail.
Integrating SmartRedis#
Typically HPC simulations are written in C, C++, Fortran or other high performance languages. Embedding the SmartRedis client usually involves compiling in the SmartRedis library into the simulation.
Because this simulation is written in Python, we can use the SmartRedis Python client to stream data to the database. To make the visualization easier, we use the SmartRedis Dataset object to hold two 2D NumPy arrays. A convenience function is provided to convert the fields into a dataset object.
# Select lines from updated simulation code highlighting
# the use of SmartRedis to stream data to another processes
from smartredis import Client, Dataset
client = Client() # Addresses passed to job through SmartSim launch
# send cylinder location only once
client.put_tensor("cylinder", cylinder.astype(np.int8))
for i in range(time_steps):
# send every 5 time_step to reduce memory consumption
if time_step % 5 == 0:
dataset = create_dataset(time_step, ux, uy)
client.put_dataset(dataset)
def create_dataset(time_step, ux, uy):
"""Create SmartRedis Dataset containing multiple NumPy arrays
to be stored at a single key within the database"""
dataset = Dataset(f"data_{time_step}")
dataset.add_tensor("ux", ux)
dataset.add_tensor("uy", uy)
return dataset
This is all the SmartRedis code needed to stream the simulation data. Note that the client does not need to have an address explicitly stated because we are going to be launching the simulation through SmartSim as shown in the cell below
[1]:
import time
import numpy as np
import matplotlib.pyplot as plt
from smartredis import Client
from smartsim import Experiment
from vishelpers import plot_lattice_vorticity, plot_lattice_norm, plot_lattice_probes
Starting the Experiment#
SmartSim, the infrastructure library, is used here to launch both the database and the simulation locally, but in separate processes. The example is designed to run on laptops, so the local launcher is used.
First the necessary libraries are imported and an Experiment instance is created. An Orchestrator database reference is intialized and launched to stage data between the simulation and this notebook where we will be performing the analysis.
[2]:
# Initialize an Experiment with the local launcher
# This will be the name of the output directory that holds
# the output from our simulation and SmartSim
exp = Experiment("finite_volume_simulation", launcher="local")
[3]:
# create an Orchestrator database reference,
# generate it's output directory, and launch it locally
db = exp.create_database(port=6780, interface="lo")
exp.generate(db, overwrite=True)
exp.start(db)
print(f"Database started at address: {db.get_address()}")
Database started at address: ['127.0.0.1:6780']
Running the Simulation#
To run the simulation, Experiment.create_run_settings is used to define how the simulation should be executed. These settings are then passed to create a reference to the simulation through a call to Experiment.create_model() which can be used to start, monitor, and stop the simulation from this notebook.
Once the model is defined it is started by passing the reference to Experiment.start() The simulation is started with the block=False argument. This runs the simulation in a nonblocking manner so that the data being streamed from the simulation can be analyzed in real time.
[4]:
# set simulation parameters we can pass as executable arguments
time_steps, seed = 3000, 42
# create "run settings" for the simulation which define how
# the simulation will be executed when passed to Experiment.start()
settings = exp.create_run_settings("python",
exe_args=["fv_sim.py",
f"--seed={seed}",
f"--steps={time_steps}"])
# Create the Model reference to our simulation and
# attach needed files to be copied, configured, or symlinked into
# the Model directory at runtime.
model = exp.create_model("fv_simulation", settings)
model.attach_generator_files(to_copy="fv_sim.py")
exp.generate(model, overwrite=True)
[5]:
# start simulation without blocking so data can be analyzed in real time
exp.start(model, block=False, summary=True)
20:36:32 HPE-C02YR4ANLVCJ SmartSim[25938:MainThread] INFO
=== Launch Summary ===
Experiment: finite_volume_simulation
Experiment Path: /home/craylabs/tutorials/online_analysis/lattice/finite_volume_simulation
Launcher: local
Models: 1
Database Status: active
=== Models ===
fv_simulation
Executable: /usr/local/anaconda3/envs/ss-py3.10/bin/python
Executable Arguments: fv_sim.py --seed=42 --steps=3000
Online Visualization#
SmartRedis is used to pull the Datasets stored in the Orchestrator database by the simulation and use matplotlib to plot the results.
In this example, we are running the visualization in an interactive manner. If instead we wanted to encapsulate this workflow to deploy on an HPC platform we could have created another Model to plot the results and launched in a similar manner to the simulation. Doing so would enable the analysis application to be executed on different resources such as GPU enabled nodes, or distributed across many nodes.
[6]:
# connect a SmartRedis client to retrieve data while the
# simulation is producing it and storing it within the
# orchestrator database
client = Client(address=db.get_address()[0], cluster=False)
# Get the cylinder location in the mesh
client.poll_key(f"cylinder", 300, 1000)
cylinder = client.get_tensor("cylinder").astype(bool)
# plot every 700th timestep
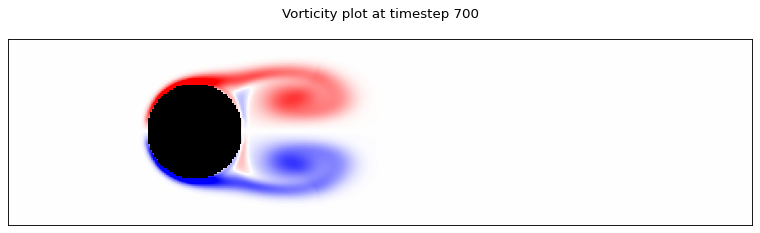
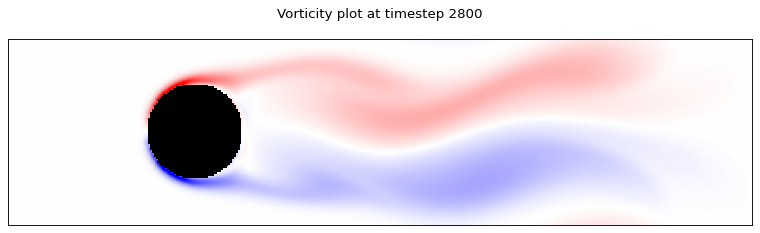
for i in range(0, time_steps, 700):
client.poll_dataset(f"data_{i}", 300, 1000)
dataset = client.get_dataset(f"data_{i}")
ux, uy = dataset.get_tensor("ux"), dataset.get_tensor("uy")
plot_lattice_vorticity(i, ux, uy, cylinder)
# Use the Experiment API to wait until the model is finished
while not exp.finished(model):
time.sleep(5)
SmartRedis Library@20-36-32:WARNING: Environment variable SR_LOG_FILE is not set. Defaulting to stdout
SmartRedis Library@20-36-32:WARNING: Environment variable SR_LOG_LEVEL is not set. Defaulting to INFO



20:37:31 HPE-C02YR4ANLVCJ SmartSim[25938:JobManager] INFO fv_simulation(26039): SmartSimStatus.STATUS_COMPLETED
Post-processing with TorchScript#
We can upload TorchScript functions to the DB. Tensors which are stored on the DB can be passed as arguments to uploaded functions and the results will be stored on the DB. This makes it possible to perform pre- and post-processing operations on tensors localli, in the DB, reducing the number of data transfers.
Uploading a script#
We can load a file containing TorchScript-compatible functionsto the DB. For example, the file ./probe.script contains the function probe_points which interpolates the values of ux and uy at some user-provided probe points. This is useful when we are interested in the value of a given fields only at specific locations.
The script looks like this:
def multi_unsqueeze(tensor, axes: List[int]):
for axis in axes:
tensor = torch.unsqueeze(tensor, axis)
return tensor
def probe_points(ux, uy, probe_x, probe_y, cylinder):
ux[cylinder>0] = 0.0
uy[cylinder>0] = 0.0
ux = multi_unsqueeze(ux, [0, 0])
uy = multi_unsqueeze(uy, [0, 0])
probe_xy = multi_unsqueeze(torch.stack((probe_x/200 - 1, probe_y/50 - 1), 2), [0])
u_probex = torch.grid_sampler(ux.double(), probe_xy.double(), 0, 0, False).squeeze()
u_probey = torch.grid_sampler(uy.double(), probe_xy.double(), 0, 0, False).squeeze()
return torch.stack((u_probex, u_probey), 2)
Note that we don’t have to import torch, as the TorchScript interpreter will recognize it as a builtin module.
We then proceed to upload the script to the DB under the key probe and add the probe points as tensors to the DB.
[7]:
client.set_script_from_file("probe", "./probe.script", device="CPU")
probe_x, probe_y = np.meshgrid(range(20, 400, 20), range(20, 100, 20))
client.put_tensor("probe_x", probe_x)
client.put_tensor("probe_y", probe_y)
Default@20-37-33:ERROR: Redis IO error when executing command: Failed to get reply: Resource temporarily unavailable
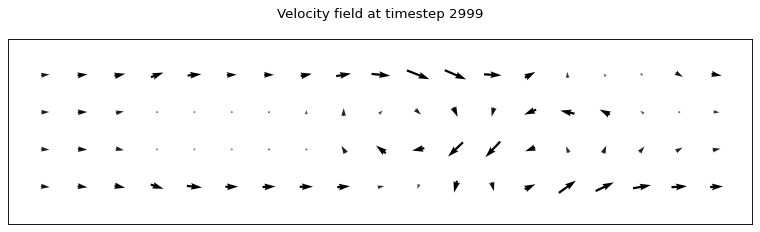
We then apply the function probe_points to the ux and uy tensors computed in the last time step of the previous simulation. Note that all tensors are already on the DB, thus we can reference them by name. Finally, we download and plot the output (a 2D velocity field), which is stored as probe_u on the DB.
[8]:
ux_name = f"{{data_{time_steps-1}}}.ux"
uy_name = f"{{data_{time_steps-1}}}.uy"
client.run_script("probe", "probe_points", inputs=[ux_name, uy_name , "probe_x", "probe_y", "cylinder"], outputs=["probe_u"])
probe_u = client.get_tensor("probe_u")
plot_lattice_probes(time_steps-1, probe_x, probe_y, probe_u)
Uploading a function inline#
In some cases, it makes sense to define the TorchScript function directly in a Python script or in a Jupyter notebook, like in this case. Let us define a simple function which computes the norm of the velocity field.
[9]:
import torch
def compute_norm(ux: torch.Tensor, uy: torch.Tensor):
return torch.sqrt(ux*ux + uy*uy)
We then store the function on the DB under the key norm_function.
[10]:
client.set_function("norm_function", compute_norm)
Note that the key we used identifies a functional unit containing the function itself: this is similar to the key used to store the probe script above. When we want to run the function, we just call it with run_script, by indicating the script key as "norm_function" and the name of the function itself as "compute_norm".
[11]:
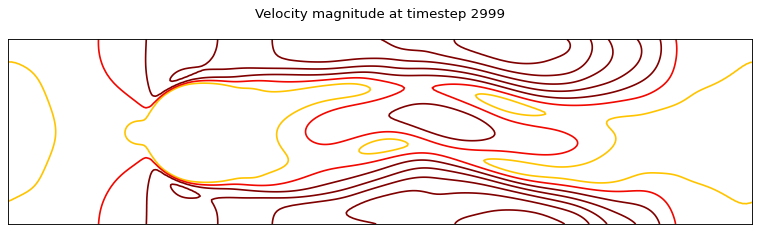
dataset = client.get_dataset(f"data_{time_steps-1}")
client.run_script("norm_function", "compute_norm", [f"{{data_{i}}}.uy", f"{{data_{i}}}.ux"], ["u"])
u = client.get_tensor("u")
plot_lattice_norm(time_steps-1, u, cylinder)
[12]:
# Optionally clear the database
client.flush_db(db.get_address())
[13]:
# terminate the database and
# release its resources
exp.stop(db)
[14]:
exp.summary(style="html")
[14]:
| Name | Entity-Type | JobID | RunID | Time | Status | Returncode | |
|---|---|---|---|---|---|---|---|
| 0 | fv_simulation | Model | 26039 | 0 | 59.2839 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 1 | orchestrator_0 | DBNode | 25963 | 0 | 75.2015 | SmartSimStatus.STATUS_CANCELLED | 0 |