Getting Started#
In this notebook, we will walk through the most basic functionalities of SmartSim.
Experiment and Models
Ensembles
Running and Communicating with the Orchestrator
Ensembles using SmartRedis
Experiments and Models#
Experiments are how users define workflows in SmartSim. The Experiment is used to create Model instances which represent applications, scripts, or generally a program. An experiment can start and stop a Model and monitor execution.
[1]:
import os
from smartsim import Experiment
The next step is to initialize an Experiment instance. The Experiment must be provided a name. This name can be any string, but it is best practice to give it a meaningful name as a broad title for what types of models the experiment will be supervising. For our purposes, our Experiment will be named "getting-started".
The Experiment also needs to have a launcher specified. Launchers provide SmartSim the ability to construct and execute complex workloads on HPC systems with schedulers (workload managers) like Slurm, or PBS. SmartSim currently supports * slurm * pbs * lsf * local (single node/laptops) * dragon * auto
If launcher="auto" is used, the experiment will attempt to find a launcher on the system, and use the first one it encounters. If a launcher cannot be found or no launcher parameter is provided, the default value of launcher="local" will be used.
For simplicity, we will start on a single host and only launch single-host jobs, and as such will set the launcher argument to "local"
[2]:
# Init Experiment and specify to launch locally
exp = Experiment(name="getting-started", launcher="local")
To run a workload through SmartSim, a Model instance must be created, and started.
Our first Model will simply print hello using the shell command echo.
Experiment.create_run_settings is used to create a RunSettings instance for our Model. RunSettings describe how a Model should be executed provided the system and available computational resources.
create_run_settings is a factory method that will instantiate a RunSettings object of the appropriate type based on the run_command argument (i.e. mpirun, aprun, srun, etc). The default argument of auto will attempt to choose a run_command based on the available system software and the launcher specified in the experiment. If run_command=None is provided, the command will be launched without one.
[3]:
# settings to execute the command "echo hello!"
settings = exp.create_run_settings(exe="echo", exe_args="hello!", run_command=None)
# create the simple model instance so we can run it.
M1 = exp.create_model(name="tutorial-model", run_settings=settings)
Once the Model has been created by the Experiment, it can be started.
By setting summary=True, we can see a summary of the experiment printed before it is launched. The summary will stay for 10 seconds, and it is useful as a last check. If we set summary=False, then the experiment would be launched immediately.
We also explicitly set block=True (even though it is the default), so that Experiment.start waits until the last Model has finished before returning: it will act like a job monitor, letting us know if processes run, complete, or fail.
[4]:
exp.start(M1, block=True, summary=True)
19:17:29 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO
=== Launch Summary ===
Experiment: getting-started
Experiment Path: /home/craylabs/tutorials/getting_started/getting-started
Launcher: local
Models: 1
Database Status: inactive
=== Models ===
tutorial-model
Executable: /bin/echo
Executable Arguments: hello!
19:17:32 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO tutorial-model(97213): SmartSimStatus.STATUS_COMPLETED
The model has completed. Let’s look at the content of the current working directory. Two files, tutorial-model.out and tutorial-model.err have been created in the Model path. To make their inspection easier, we can define a helper function.
[5]:
def get_files(model):
"""Get output and error file of a Model"""
outputfile = os.path.join(model.path, model.name+".out")
errorfile = os.path.join(model.path, model.name+".err")
return outputfile, errorfile
outputfile, errorfile = get_files(M1)
print("Content of tutorial-model.out:")
with open(outputfile, 'r') as fin:
print(fin.read())
print("Content of tutorial-model.err:")
with open(errorfile, 'r') as fin:
print(fin.read())
Content of tutorial-model.out:
hello!
Content of tutorial-model.err:
The .out file contains the output generated by tutorial-model, and the .err file would contain the error messages generated by it. Since there were no errors, the .err file is empty.
Now let’s run two different Model instances at the same time. This is just as easy as running one Model, and takes the same steps. This time, we will skip the summary.
[6]:
run_settings_1 = exp.create_run_settings(exe="echo", exe_args="hello!", run_command=None)
run_settings_2 = exp.create_run_settings(exe="sleep", exe_args="5", run_command=None)
model_1 = exp.create_model("tutorial-model-1", run_settings_1)
model_2 = exp.create_model("tutorial-model-2", run_settings_2)
exp.start(model_1, model_2)
19:17:37 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO tutorial-model-1(97239): SmartSimStatus.STATUS_COMPLETED
19:17:40 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO tutorial-model-2(97250): SmartSimStatus.STATUS_RUNNING
19:17:41 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO tutorial-model-2(97250): SmartSimStatus.STATUS_COMPLETED
For users of parallel applications, launch binaries (run commands) can also be specified in RunSettings. For example, if mpirun is installed on the system, we can run a model through it, by specifying it as run_command in create_run_settings.
Please note that to run this you need to have OpenMPI installed.
[7]:
# settings to execute the command "mpirun -np 2 echo hello world!"
openmpi_settings = exp.create_run_settings(exe="echo",
exe_args="hello world!",
run_command="mpirun")
openmpi_settings.set_tasks(2)
# create and start the MPI model
ompi_model = exp.create_model("tutorial-model-mpirun", openmpi_settings)
exp.start(ompi_model, summary=True)
19:17:45 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO
=== Launch Summary ===
Experiment: getting-started
Experiment Path: /home/craylabs/tutorials/getting_started/getting-started
Launcher: local
Models: 1
Database Status: inactive
=== Models ===
tutorial-model-mpirun
Executable: /bin/echo
Executable Arguments: hello world!
Run Command: /usr/local/bin/mpirun
Run Arguments:
n = 2
19:17:47 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO tutorial-model-mpirun(97310): SmartSimStatus.STATUS_COMPLETED
This time, since we asked mpirun to run two tasks by calling openmpi_settings.set_tasks(2), in the output file we should find the line hello world! twice.
[8]:
outputfile, _ = get_files(ompi_model)
print("Content of tutorial-model-mpirun.out:")
with open(outputfile, 'r') as fin:
print(fin.read())
Content of tutorial-model-mpirun.out:
hello world!
hello world!
Ensembles#
In the previous example, the two Model instances were created separately. The Ensemble SmartSim object is a more convenient way of setting up multiple models, potentially with different configurations. Ensembles are groups of Model instances that can be treated as a single reference. We start by specifying RunSettings similar to how we did with our Models.
[9]:
# define how we want each ensemble member to execute
# in this case we create settings to execute "sleep 3"
ens_settings = exp.create_run_settings(exe="sleep", exe_args="3")
Then, instead of creating a single model like we did in previously, we will call the Experiment.create_ensemble method to create an Ensemble. Let’s assume we want to run the same experiment four times in parallel. We will then pass the method the same arguemnts that we might pass Experiment.create_model in addition to the replicas=4 argument. Finally, we simply start the Ensemble the same way we wold start a Model.
[10]:
ensemble = exp.create_ensemble("ensemble-replica",
replicas=4,
run_settings=ens_settings)
exp.start(ensemble, summary=True)
19:17:50 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO
=== Launch Summary ===
Experiment: getting-started
Experiment Path: /home/craylabs/tutorials/getting_started/getting-started
Launcher: local
Database Status: inactive
=== Ensembles ===
ensemble-replica
Members: 4
Batch Launch: None
19:17:55 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble-replica_0(97347): SmartSimStatus.STATUS_COMPLETED
19:17:56 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO ensemble-replica_1(97348): SmartSimStatus.STATUS_COMPLETED
19:17:56 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO ensemble-replica_2(97349): SmartSimStatus.STATUS_COMPLETED
19:17:56 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO ensemble-replica_3(97350): SmartSimStatus.STATUS_COMPLETED
19:17:57 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble-replica_1(97348): SmartSimStatus.STATUS_COMPLETED
19:17:57 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble-replica_2(97349): SmartSimStatus.STATUS_COMPLETED
19:17:57 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble-replica_3(97350): SmartSimStatus.STATUS_COMPLETED
From the output, we see that four copies of our Model, named ensemble-replica_0, ensemble-replica_1, … were run. In each output file, we will see that the same output was generated.
Now let’s imagine that we don’t want to run the same model four times, but we want to run variations of it. One way of doing this would be to define four models, and starting them through the Experiment.
For a few, simple Models, this would be OK, but what if we needed to run a large number of models, which only differ for some parameter? Defining and adding each one separately would be tedious. For such cases, we will rely on a parameterized Ensemble of models.
Say we had a python file output_my_parameter.py with this contents:
# contents of output_my_parameter.py
import time
time.sleep(2)
print("Hello, my name is ;tutorial_name; " +
"and my parameter is ;tutorial_parameter;")
Our goal is to run
python output_my_parameter.py
with multiple parameter values substituted where the text contains ;tutorial_name; and ;tutorial_parameter;. Clearly, we could pass the parameters as arguments, but in some cases, this could not be possible (e.g. if the parameters were stored in a file or the executable would not accept them from the command line).
First thing first, is that we must again create our run settings:
[11]:
rs = exp.create_run_settings(exe="python", exe_args="output_my_parameter.py")
Then, we define the parameters we are going to set and values for those parameters in a dictionary. In this example, we are setting:
tutorial_namewith values"Ellie"and"John"tutorial_parameterwith values2and11
In the original file output_my_parameter.py, which acts as a template, they occur as ;tutorial_name; and ;tutorial_parameter;. The semi-colons are used to perform a regexp substitution with the desired values. The semi-colon in this case, is called a tag and can be changed.
We pass the parameter ditionary to Experiment.create_ensemble, along with the argument perm_strategy="all_perm". This argument means that we want all possible permutations of the given parameters, which are stored in the argument params. We have two options for both parameters, thus our ensemble will run 4 instances of the same Experiment, just using a different copy of output_my_parameter.py created by calling Experiment.generate(). We attach the template file to the
Ensemble instance, generate the augmented python files, and run the experiment.
[12]:
params = {
"tutorial_name": ["Ellie", "John"],
"tutorial_parameter": [2, 11]
}
ensemble = exp.create_ensemble("ensemble", params=params, run_settings=rs, perm_strategy="all_perm")
# to_configure specifies that the files attached should be read and tags should be looked for
config_file = "./output_my_parameter.py"
ensemble.attach_generator_files(to_configure=config_file)
exp.generate(ensemble, overwrite=True)
exp.start(ensemble)
19:18:06 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_0(97408): SmartSimStatus.STATUS_COMPLETED
19:18:06 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_1(97409): SmartSimStatus.STATUS_COMPLETED
19:18:06 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_3(97421): SmartSimStatus.STATUS_COMPLETED
19:18:07 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO ensemble_2(97410): SmartSimStatus.STATUS_COMPLETED
19:18:08 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_2(97410): SmartSimStatus.STATUS_COMPLETED
We can see from the output that four instances of our experiment were run, each one named like the Experiment, with a numeric suffix at the end: ensemble_0, ensemble_1, etc. The call to Experiment.generate() created isolated output directories for each created Model in the ensemble and each ensemble member generated its own output files, which was stored in its respective directory.
[13]:
for id in range(4):
outputfile, _ = get_files(ensemble.entities[id])
print(f"Content of {outputfile}:")
with open(outputfile, 'r') as fin:
print(fin.read())
Content of /home/craylabs/tutorials/getting_started/getting-started/ensemble/ensemble_0/ensemble_0.out:
Hello, my name is Ellie and my parameter is 2
Content of /home/craylabs/tutorials/getting_started/getting-started/ensemble/ensemble_1/ensemble_1.out:
Hello, my name is Ellie and my parameter is 11
Content of /home/craylabs/tutorials/getting_started/getting-started/ensemble/ensemble_2/ensemble_2.out:
Hello, my name is John and my parameter is 2
Content of /home/craylabs/tutorials/getting_started/getting-started/ensemble/ensemble_3/ensemble_3.out:
Hello, my name is John and my parameter is 11
That’s it! All possible permutations of the input parameters were used to execute the experiment! Sometimes, the parameter space can be too large to be explored exhaustively. In that case, we can use a different permutation strategy, i.e. random. For example, if we want to only use two possible random combinations of our parameter space, we can run the following code, where we specify n_models=2 and perm_strategy="random".
[14]:
params = {
"tutorial_name": ["Ellie", "John"],
"tutorial_parameter": [2, 11]
}
ensemble = exp.create_ensemble("param_ensemble", params=params, run_settings=rs, perm_strategy="random", n_models=2)
config_file = "./output_my_parameter.py"
ensemble.attach_generator_files(to_configure=config_file)
exp.generate(ensemble, overwrite=True)
exp.start(ensemble)
19:18:17 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO param_ensemble_0(97484): SmartSimStatus.STATUS_COMPLETED
19:18:17 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO param_ensemble_1(97495): SmartSimStatus.STATUS_COMPLETED
Another possible permutation strategy is stepped, but it is also possible to pass a function, which will need to generate combinations of parameters starting from the dictionary. Please refer to the documentation to learn more about this.
It is also possible to use different delimiters for the parameter regexp. For example, if we had simmlarly parameterized file named output_my_parameter_new_tag.py, with contents:
# Contents of output_my_parameter_new_tag.py
import time
time.sleep(2)
print("Hello, my name is @tutorial_name@ " +
"and my parameter is @tutorial_parameter@")
We would want to use @, instead of ;, as our tag. We can trivially make this adaptation by passing a tag argument to our Experiment.generate call.
[15]:
rs = exp.create_run_settings(exe="python", exe_args="output_my_parameter_new_tag.py")
params = {
"tutorial_name": ["Ellie", "John"],
"tutorial_parameter": [2, 11]
}
ensemble = exp.create_ensemble("ensemble_new_tag",
params=params,
run_settings=rs,
perm_strategy="all_perm")
config_file = "./output_my_parameter_new_tag.py"
ensemble.attach_generator_files(to_configure=config_file)
exp.generate(ensemble, overwrite=True, tag='@')
exp.start(ensemble)
19:18:23 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_new_tag_0(97520): SmartSimStatus.STATUS_COMPLETED
19:18:23 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_new_tag_1(97521): SmartSimStatus.STATUS_COMPLETED
19:18:23 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_new_tag_3(97523): SmartSimStatus.STATUS_COMPLETED
19:18:24 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO ensemble_new_tag_2(97522): SmartSimStatus.STATUS_COMPLETED
19:18:25 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO ensemble_new_tag_2(97522): SmartSimStatus.STATUS_COMPLETED
Last, we can see all the kernels we have executed by calling Experiment.summary()
[16]:
print(exp.summary())
| | Name | Entity-Type | JobID | RunID | Time | Status | Returncode |
|----|-----------------------|---------------|---------|---------|--------|---------------------------------|--------------|
| 0 | tutorial-model | Model | 97213 | 0 | 2.0073 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 1 | tutorial-model-1 | Model | 97239 | 0 | 2.2181 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 2 | tutorial-model-2 | Model | 97250 | 0 | 6.0111 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 3 | tutorial-model-mpirun | Model | 97310 | 0 | 2.0072 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 4 | ensemble-replica_0 | Model | 97347 | 0 | 4.6530 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 5 | ensemble-replica_1 | Model | 97348 | 0 | 6.4457 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 6 | ensemble-replica_2 | Model | 97349 | 0 | 6.2330 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 7 | ensemble-replica_3 | Model | 97350 | 0 | 6.0211 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 8 | ensemble_0 | Model | 97408 | 0 | 4.6442 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 9 | ensemble_1 | Model | 97409 | 0 | 4.4313 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 10 | ensemble_3 | Model | 97421 | 0 | 4.0064 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 11 | ensemble_2 | Model | 97410 | 0 | 6.2264 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 12 | param_ensemble_0 | Model | 97484 | 0 | 4.2159 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 13 | param_ensemble_1 | Model | 97495 | 0 | 4.0068 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 14 | ensemble_new_tag_0 | Model | 97520 | 0 | 4.6525 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 15 | ensemble_new_tag_1 | Model | 97521 | 0 | 4.4403 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 16 | ensemble_new_tag_3 | Model | 97523 | 0 | 4.0074 | SmartSimStatus.STATUS_COMPLETED | 0 |
| 17 | ensemble_new_tag_2 | Model | 97522 | 0 | 6.2288 | SmartSimStatus.STATUS_COMPLETED | 0 |
Orchestrator#
The Orchestrator is an in-memory database (Redis/KeyDB) that is launched prior to all other entities within an Experiment. The Orchestrator can be used to store and retrieve data across languages (Fortran, C, C++, Python) during the course of an experiment and across multiple workloads. In order to stream data into or receive data from the Orchestrator, one of the SmartSim clients (SmartRedis) has to be used within your workload.
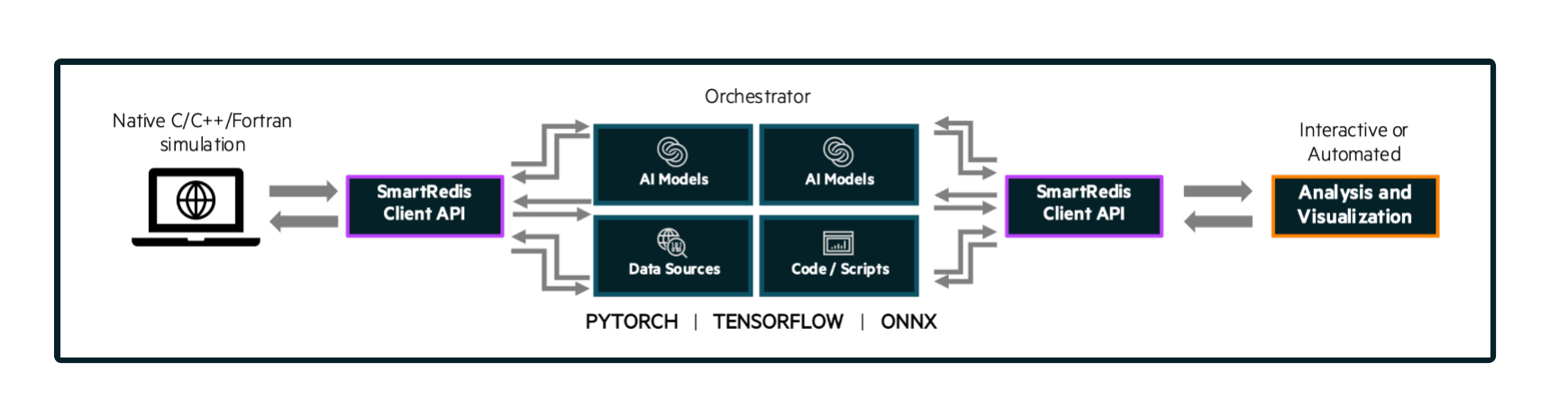
The Orchestrator is capable of hosting and executing AI models written in Python on CPU or GPU. The Orchestrator supports models written with TensorFlow, Pytorch, or models saved in an ONNX format (e.g. scikit-learn). See the inference tutorial for more information on how to use the machine learning runtimes built into the Orchestrator database.
Orchestrators can either be deployed on a single host, or many hosts as shown in the diagram below.
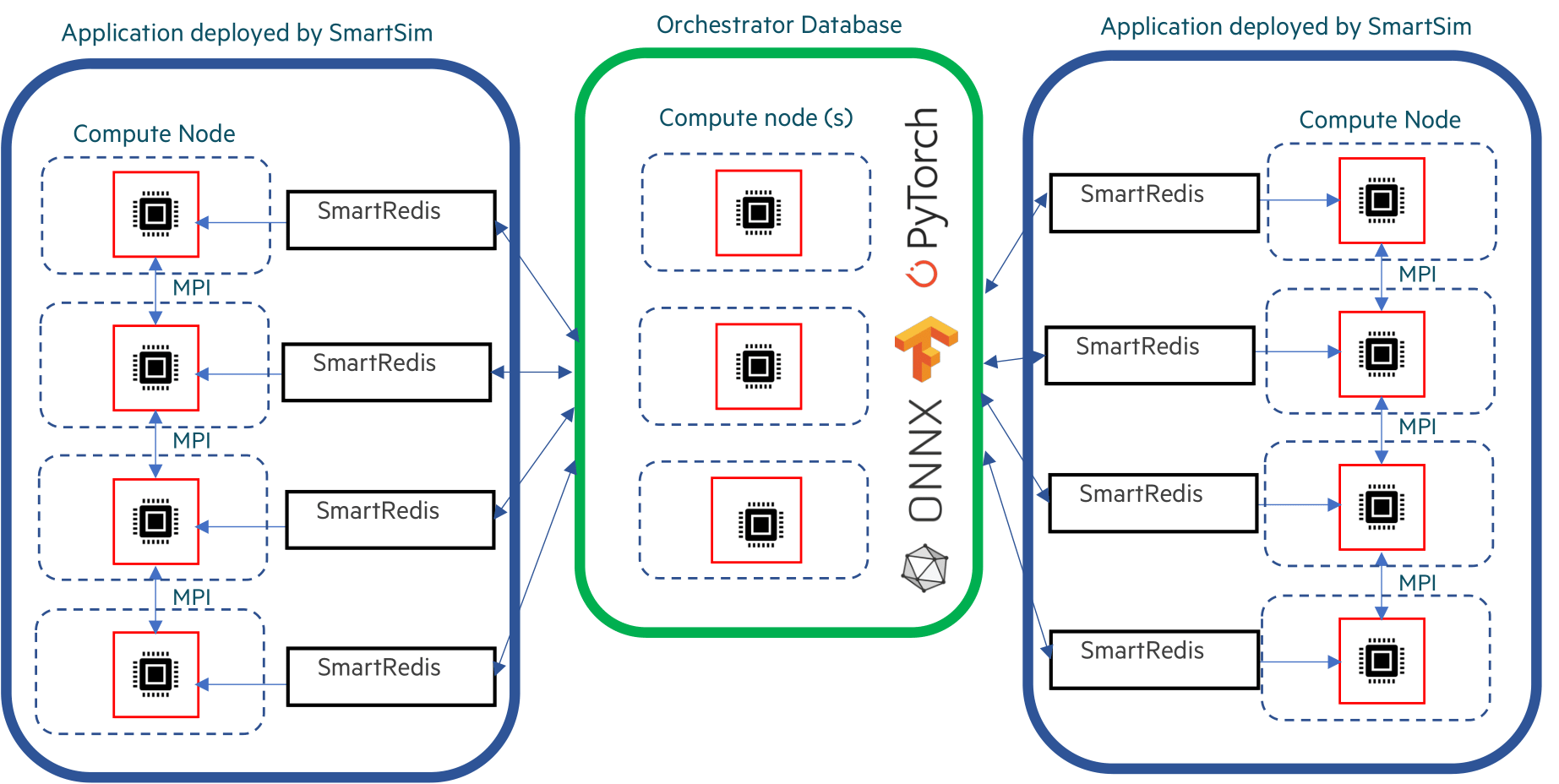
In this tutorial, a single-host host Orchestrator is deployed locally (as we specified local for the Experiment launcher) and used to demonstrate how to use the SmartRedis Python client within a workload.
[17]:
from smartredis import Client
import numpy as np
REDIS_PORT=6899
[18]:
# start a new Experiment for this section
exp = Experiment("tutorial-smartredis", launcher="local")
# create and start an instance of the Orchestrator database
db = exp.create_database(db_nodes=1,
port=REDIS_PORT,
interface="lo")
# create an output directory for the database log files
exp.generate(db)
# start the database
exp.start(db)
Now that the Orchestrator is running, a SmartRedis client can be used to store and retrieve data from the database.
[19]:
# connect a SmartRedis client at the address supplied by the launched
# Orchestrator instance.
# Cluster=False as the Orchestrator was deployed on a single compute host (local)
client = Client(address=db.get_address()[0], cluster=False)
SmartRedis Library@19-18-39:WARNING: Environment variable SR_LOG_FILE is not set. Defaulting to stdout
SmartRedis Library@19-18-39:WARNING: Environment variable SR_LOG_LEVEL is not set. Defaulting to INFO
Then, we can use the client to put and retrieve Tensors. Tensors are the native array format of the client language being used. For example, in Python, NumPy arrays are the Tensor format for SmartRedis. Each stored tensors needs a unique key at which to be stored.
[20]:
send_tensor = np.ones((4,3,3))
client.put_tensor("tutorial_tensor_1", send_tensor)
receive_tensor = client.get_tensor("tutorial_tensor_1")
print('Receive tensor:\n\n', receive_tensor)
Receive tensor:
[[[1. 1. 1.]
[1. 1. 1.]
[1. 1. 1.]]
[[1. 1. 1.]
[1. 1. 1.]
[1. 1. 1.]]
[[1. 1. 1.]
[1. 1. 1.]
[1. 1. 1.]]
[[1. 1. 1.]
[1. 1. 1.]
[1. 1. 1.]]]
With the SmartRedis Client and its possible to store and run a PyTorch, TensorFlow, or ONNX model in the database. The example below shows a PyTorch model being created, set in the database, and called from a SmartRedis client.
For more information on ML inference in SmartSim, see the inference tutorial.
[21]:
import torch
import torch.nn as nn
# taken from https://pytorch.org/docs/master/generated/torch.jit.trace.html
class Net(nn.Module):
def __init__(self):
super().__init__()
self.conv = nn.Conv2d(1, 1, 3)
def forward(self, x):
return self.conv(x)
net = Net()
example_forward_input = torch.rand(1, 1, 3, 3)
module = torch.jit.trace(net, example_forward_input)
# Save the traced model to a file
torch.jit.save(module, "./torch_cnn.pt")
Now we send the model to the database, again, we assign it a unique key, tutorial-cnn. This key is provided to run the model in the Client.run_model method.
[22]:
# Set the model in the Redis database from the file
client.set_model_from_file("tutorial-cnn", "./torch_cnn.pt", "TORCH", "CPU")
Default@19-18-41:ERROR: Redis IO error when executing command: Failed to get reply: Resource temporarily unavailable
[23]:
# Put a tensor in the database as a test input
data = torch.rand(1, 1, 3, 3).numpy()
client.put_tensor("torch_cnn_input", data)
# Run model and retrieve the output
client.run_model("tutorial-cnn", inputs=["torch_cnn_input"], outputs=["torch_cnn_output"])
out_data = client.get_tensor("torch_cnn_output")
Notice that we could have defined the model as an object (without storing it on disk) and send it to the DB using set_model instead of set_model_from_file. We can do the same thing for any Python function. For example, let’s define a simple function takes a NumPy tensor as input.
[24]:
def max_of_tensor(array):
"""Sample torchscript script that returns the
highest element in an array.
"""
# return the highest element
return array.max(1)[0]
sample_array_1 = np.array([np.arange(9.)])
print(sample_array_1)
print("Max:")
print(max_of_tensor(sample_array_1))
[[0. 1. 2. 3. 4. 5. 6. 7. 8.]]
Max:
8.0
Now let’s store this function so it can be called, assigning it the key max-of-tensor:
[25]:
client.set_function("max-of-tensor", max_of_tensor)
A Client can now be used to call this function where it will run wherever the database is deployed (on CPU or GPU)
[26]:
client.put_tensor("script-data-1", sample_array_1)
client.run_script(
"max-of-tensor", # key of our script
"max_of_tensor", # function to be called
["script-data-1"],
["script-output"],
)
out = client.get_tensor("script-output")
print(out)
[8.]
And, as expected, we obtain the same result we obtained when we ran the function locally. To clean up, we need to tear down the DB. We do this by stopping the Orchestrator.
[27]:
exp.stop(db)
Ensembles using SmartRedis#
In Section 1.2 we used Ensembles. What would happen if Models which are part of an Ensemble tried to put their tensors on the DB using SmartRedis? Unless we used unique keys across the running programs, several tensors (or objects) would have the same key, and this key collision would result in unexpected behavior. In other words, if in the source code of one program, a tensor with key tensor1 was put on the DB, then each replica of the program would put a tensor with the key
tensor1. SmartSim and SmartRedis can avoid key collision by prepending program-unique prefixes to Model workloads launched through SmartSim.
Instead of creating a new Experiment for this section, we will use the previous Experiment and relaunch the Orchestrator using the db reference that is already defined.
[28]:
exp.start(db)
Now let’s add two replicas of the same Model. Basically, it is a simple producer, which puts a tensor on the DB. The code for it is in producer.py.
[29]:
rs_prod = exp.create_run_settings("python", f"producer.py --redis-port {REDIS_PORT}")
ensemble = exp.create_ensemble(name="producer",
replicas=2,
run_settings=rs_prod)
We add a consumer, which will just retrieve the tensors put by the two producers and check that they are what it expects.
[30]:
rs_consumer = exp.create_run_settings("python", f"consumer.py --redis-port {REDIS_PORT}")
consumer = exp.create_model("consumer", run_settings=rs_consumer)
We need to register incoming entities, i.e. entities for which the prefix will have to be known by other entities. When we will start the Experiment, environment variables will be set to let all entities know which incoming entities are present.
[31]:
consumer.register_incoming_entity(ensemble.models[0])
consumer.register_incoming_entity(ensemble.models[1])
Finally, we attach the files to the experiments, generate them, and run!
[32]:
ensemble.attach_generator_files(to_copy=['producer.py'])
consumer.attach_generator_files(to_copy=['consumer.py'])
exp.generate(ensemble, overwrite=True)
exp.generate(consumer, overwrite=True)
# start the models
exp.start(ensemble, consumer, summary=True)
19:18:53 HPE-C02YR4ANLVCJ SmartSim[97173:MainThread] INFO
=== Launch Summary ===
Experiment: tutorial-smartredis
Experiment Path: /home/craylabs/tutorials/getting_started/tutorial-smartredis
Launcher: local
Models: 1
Database Status: active
=== Ensembles ===
producer
Members: 2
Batch Launch: None
=== Models ===
consumer
Executable: /usr/local/anaconda3/envs/ss-py3.10/bin/python
Executable Arguments: consumer.py --redis-port 6899
19:18:58 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO producer_0(97711): SmartSimStatus.STATUS_COMPLETED
19:18:58 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO producer_1(97712): SmartSimStatus.STATUS_COMPLETED
19:18:58 HPE-C02YR4ANLVCJ SmartSim[97173:JobManager] INFO consumer(97713): SmartSimStatus.STATUS_COMPLETED
The producers produced random NumPy tensors, and we can see that the consumer was able to retrieve both of them from the DB, by looking at its output.
[33]:
outputfile, _ = get_files(consumer)
with open(outputfile, 'r') as fin:
print(fin.read())
SmartRedis Library@19-18-54:WARNING: Environment variable SR_LOG_FILE is not set. Defaulting to stdout
SmartRedis Library@19-18-54:WARNING: Environment variable SR_LOG_LEVEL is not set. Defaulting to INFO
Tensor for producer_0 is: [[[[0.40963388 0.66147363 0.88239209]
[0.67788696 0.66730329 0.26504813]
[0.80848382 0.96430444 0.75951969]]]]
Tensor for producer_1 is: [[[[0.67515573 0.28582205 0.79349604]
[0.78848592 0.67902375 0.54826283]
[0.01769311 0.55995054 0.47818324]]]]
As usual, let’s shutdown the DB, by stopping the Orchestrator.
[34]:
exp.stop(db)